We work at the spatial biology-technology interface to understand chromatin<–>synapse communication in the mammalian brain.
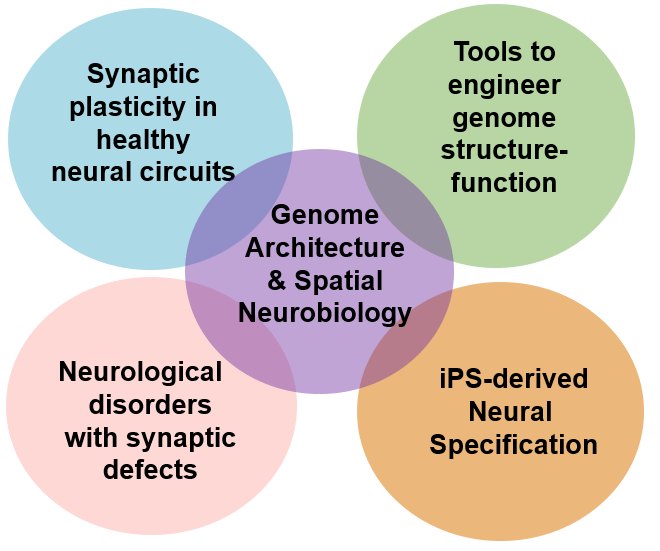
The Cremins lab aims to understand how chromatin works through long-range physical folding mechanisms to encode neuronal specification and long-term synaptic plasticity in healthy and diseased neural circuits. We pursue a multi-disciplinary approach integrating data across biological scales in the brain, including molecular Chromosome-Conformation-Capture sequencing technologies, single-cell imaging, optogenetics, genome engineering, induced pluripotent stem cell differentiation to neurons/organoids, and in vitro and in vivo electrophysiological measurements.
-
Dr. Chuanbin Su posts a first-author preprint on the development of his new technology: MASTR-seq for Multiplexed Analysis of Short Tandem Repeats with Sequencing. Congratulations Chuanbin!https://www.biorxiv.org/content/10.1101/2024.04.29.591790v1
-
Harshini Chandrashekar presents her work at the Genetics department work in progress talks.
-
Our alum, Dr. Ji Hun Kim, wins his first research grant as an assistant professor at KAIST. Dr. Kim is using the LADL (Light-activated Dynamic Looping) technology he built to study the role for loops in gene expression regulation in cancer. Congratulations Ji Hun!
-
Our alum, Dr. Thomas Gilgenast, visits KAIST to visit Ji Hun and give a seminar on his collaborative work with Scharam Akbarian. Congratulations Thomas!
-
Congratulations to Harshini Chandrashekar for winning first prize for her oral presentation in the Bioengineering PhD program symposium.
-
Warm congratulations to Harshini, Zoltan, Heesun, Avi, Ali, and Han-Seul for posting their new BioRxiv preprint uncovering a role for dynamic multi-loops in gene expression dysregulation in post-mitotic human neurons homozygously-engineered with familial Alzheimer’s disease mutations. https://www.biorxiv.org/content/10.1101/2024.02.27.582395v1
-
Congratulations to Dr. Peibo Xu – one of his first-author PhD papers entitled ‘High-throughput mapping of single-neuron projection and molecular features by retrograde barcoded labeling‘ is now published in eLife. Congratulations Peibo!https://pubmed.ncbi.nlm.nih.gov/38390967/
-
MSTP student Han-Seul Ryu passes her proposal and becomes a PhD candidate. Congratulations Han-Seul!
-
Huge congratulations to MSTP student Kenneth Pham on receiving his first NIH Notice of Award for his F30 fellowship grant. Great job Kenneth!
-
Edda Schulz writes a commentary on our team’s fragile x paper in Molecular Cell. https://www.sciencedirect.com/science/article/pii/S1097276524000017?dgcid=author
-
NIMH writes a news article on our Fragile X Syndrome work: https://www.nimh.nih.gov/news/science-news/2024/researchers-expand-understanding-of-genetic-mechanisms-underlying-fragile-x-syndrome
-
Kenneth Pham receives a fundable score on his NIH NRSA F30 proposal
Our work is supported by the New York Stem Cell Foundation, the Alfred P. Sloan Foundation, the National Science Foundation, the National Institute of Mental Health, the National Institute of Neural Disorders and Stroke, NIH Common fund initiatives, the Friedreich’s Ataxia Research Alliance, the Chan Zuckerberg Initiative, and the NIH 4D Nucleome Common Fund Initiative.

